Batch
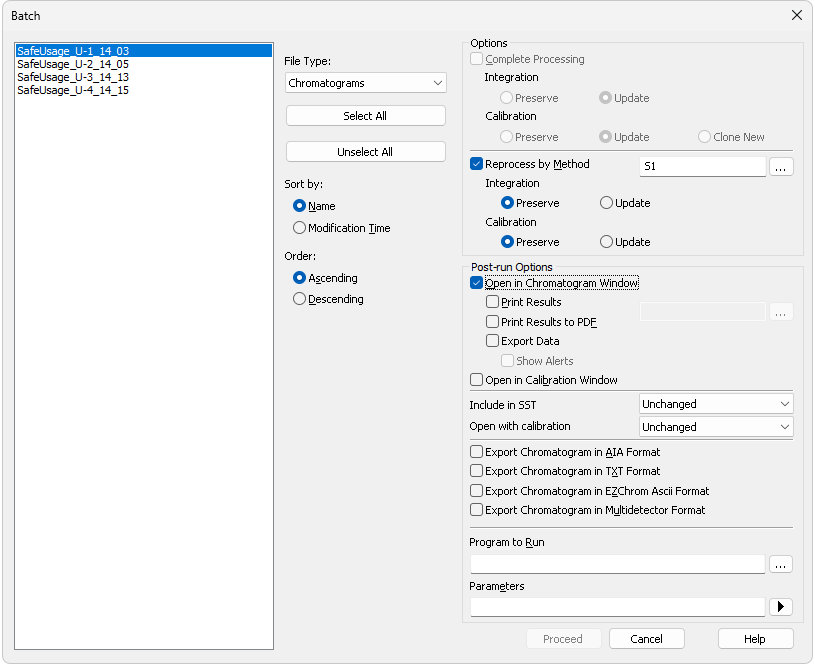
Batch processes single chromatograms, automatically and simultaneously executes selected actions with numerous chromatograms or reprocesses whole sequences. When invoked, the Batch dialog will be displayed.
Batch
Lists all chromatograms or sequences from the current project according to the settings in the File Type field. Left click the mouse while holding the Shift or Ctrl key to select and deselect multiple files. This will enable you to progressively mark any arbitrary number of displayed files.
Selects the type of files to be batch processed.
Chromatograms - The list will display all chromatograms from the data subdirectory and its subdirectories.
Calibration Standards - The list will display all chromatograms from the calibration subdirectory and its subdirectories. (i.e. calibration standards).
Sequence Files - Sequences from the current project will be displayed. You can change information on several columns (Run, Sample Type, Lvl, Report Style and Post-run Options) of the already measured sequence file to define which rows should be reprocessed and how.
Note:
Each sequence file remembers chromatograms, which were measured last and, therefore, it is not necessary to update the File Name column. Chromatograms not found are ignored.
Caution:
When a sequence is run repeatedly, the list of measured files in that sequence will be reset and the original files cannot be processed using the Batch command.
Quickly selects or deselects all files in the list.
Sorts files by Name or by the Time upon which they were last saved.
Sorts files in Ascending or Descending order based on Sort by setting. As files are processed in the order in which they appear in the list, the order of processing can be changed this way.
Options section
Options section is divided to two halves - one for Complete Processing, the other for Reprocess by Method. These sections are mutually exclusive.
This command is active only for processing chromatograms according to the sequence file (the File Type field must be set to Sequence Files). In this case, all chromatograms will be processed according to the current sequence table contents and methods used, including the eventual recalibration. This means that you can batch change headers of already measured chromatograms and reintegrate them, if you modify the integration parameters in used methods, etc.
Note:
The Post-run Options section is disabled as the post-run activities will be performed exactly according to the sequence settings on the particular sequence row during the reprocessing.
Caution:
Only the last set of measured chromatograms will be processed if the same sequence was used more than once and reset in between uses.
Governs the behavior of the integration tables contained within chromatograms that originated from the given sequence. With Preserve option the Integration tables will not be changed during the reprocess, with Update option (default) the Integration tables of chromatograms will be replaced by the integration tables of signals as they are in the method used on a given sequence table row.
Governs the behavior of calibration handling in the sequence being reprocessed. With Preserve option the calibrations connected to the particular chromatograms will be left as they are (see note below), for sequence rows with calibration standards and calibration level filled in the calibration will not be recalibrated. With Update option (default), the calibrations that are currently connected to the chromatograms will be used for potential recalibration (if the sequence contains samples marked as calibration standards with level filled in) and the latest version of the calibration available at that moment will be connected to the chromatogram being reprocessed. In case the sequence used calibration cloning, the original clone will be used. With the Clone New option, in case the sequence used calibration cloning, a new set of calibrations will be cloned, calibrated and the chromatograms will be reconnected to these new clones. In case the sequence does not use calibration cloning, the option behaves same as Update option.
Note:
The Open with stored calibration checkbox in the Sequence controls how the chromatogram is opened - if checked, it is opened with the version of the calibration last stored in the chromatogram (in case of Update, the version after reprocessing), and any subsequent changes do not affect it. Otherwise, the chromatogram opens with the linked "live" calibration - any calibration change is immediately propagated to the chromatogram results.
Reprocesses all selected chromatograms according to the selected method. By default, displayed method matches method in the Single Analysis dialog. This option can be used with both chromatograms and sequences - in case of sequences, selected sequence only serves as a source of selection of the used chromatograms, e.g. no recalibration will be performed no matter the settings.
Integration
Governs the behavior of the integration tables contained within selected chromatograms. With Preserve option the Integration tables will not be changed during the reprocess, with Update option (default) the Integration tables of chromatograms will be replaced by the integration tables of signals in the method used for reprocessing.
Calibration
Governs the behavior of calibration handling in the chromatograms being reprocessed. With Preserve option the calibrations connected to the particular chromatograms will be left as they are (see note below), with Update option (default), the calibrations in the chromatograms will be replaced by the calibration set in the method used for reprocessing.
Note:
The Open with Calibration drop-down list controls how the chromatogram is opened, when Unchanged is selected, the setting in the chromatogram itself is used. If Stored, it is opened with the version of the calibration last stored in the chromatogram (in case of Update, the version after reprocessing), and any subsequent changes do not affect it. Otherwise, the chromatogram opens with the linked "live" calibration - any calibration change is immediately propagated to the chromatogram results.
This section allows to perform the post-run actions same as if set in Single Analysis dialog or Sequence window. The actions will be carried out on selected chromatograms or chromatograms measured from selected sequence. The actions can be performed standalone or in a combination with Reprocess by Method option.
Chromatogram will be opened in the Chromatogram window. Opening of the chromatogram in Chromatogram window is necessary for several further actions (printing or exporting result tables) so these further options are available only if the Open in Chromatogram Window checkbox is enabled. Chromatograms are opened in the window according to the current display options there (Overlay mode on/off, number of chromatograms opened at the same time, table layouts) which may influence printed results (e.g. contents of a printed Summary table).
Prints a report according to the selected report style. Each report style is adjustable in the Report Setup dialog which is accessible from different windows and dialogs, such as Single Analysis and Method Setup dialogs, Sequence, Chromatogram and Calibration windows and others.
Prints a report to the *.PDF file according to the selected report style.
Each chromatogram will be exported according to the settings in the Export Data dialog.
Sets the display of system alerts possibly appearing during the export - if checked, the alerts are displayed in user interface in addition to saving them in the log (which happens no matter the setting).
Chromatogram will be opened in the Calibration window, no further actions are performed with it.
Selects whether the batch-processed chromatograms will be included in SST calculations. Available options are Exclude, Include and Unchanged.
Selects the type of calibration with which the chromatogram will be opened. Available options are Linked, Stored or Unchanged. Linked and Stored values influence how the chromatogram will be opened for particular other actions set in the Batch dialog (such as print or export) and are then saved to the chromatogram itself, so that after the reprocess the chromatogram will be opened by default from the file manager in the same manner as well. Unchanged value leaves the same parameter in the chromatogram untouched.
Note:
The setting also influences calibration handling in case Preserve option is selected in Reprocess by Method section - with chromatograms opened using the Stored option the calibration will stay exactly as it was before reprocessing, with Linked option the latest version of the same calibration will be used within chromatogram.
Export Chromatogram in AIA Format
Exports a chromatogram in AIA format. Corresponds to the File - Export Chromatogram command in the Chromatogram window.
The CDF format file will have the same name as the chromatogram and will be saved in the same (data or calibration) subdirectory of the current project as the exported chromatogram.
Export Chromatogram in TXT Format
Exports a chromatogram in TXT format. Corresponds to the File - Export Chromatogram command in the Chromatogram window.
The TXT format file will have the same name as the chromatogram and will be saved in the same (data or calibration) subdirectory of the current project as the exported chromatogram.
Export Chromatogram in EZChrom Ascii Format
Exports a chromatogram in ASC text format, as it is used by the EZ-Chrom program. Corresponds to the File - Export Chromatogram command in the Chromatogram window.
A file with the ASC suffix will have the same name as the chromatogram and will be saved in the same (data or calibration) subdirectory of the current project as the exported chromatogram.
Export Chromatogram in Multidetector Format
Exports a chromatogram in CHR text format so that all signals of multidetector chromatogram are exported to a single file.
A file with the CHR suffix will have the same name as the chromatogram and will be saved in the same (data or calibration) subdirectory of the current project as the exported chromatogram.
Allows to select the program that should be started as part of the chromatogram reprocess (e.g. EXCEL.EXE).

Searches for the program to be run. Standard MS Windows window will be displayed for program search.
Sets the parameters of an initiated program; these parameters are generally entered in the command line after the program name (e.g. name of loaded file, etc.).
The following variables can be used in the Parameters field (the  button allows to select them interactively):
button allows to select them interactively):
- %f will be replaced by the name of the chromatogram after export.
- %e will be replaced by the name of the file after export. This does not apply when exporting multidetector chromatograms in the .TXT or .AIA formats.
Runs batch processing.