Reprocessing whole sequence can be convenient if performing reintegration, recalibration, or changing types of samples is needed. It can also be used with calibration cloning, more in the topic Complete processing with calibration cloning. Steps below described process where sequeunce is already measured by template method with optimized integration parameters, for more see Save Method as a Template.
- Measure the sequence with no post run options set and/or no Levels for standards filled in (Typically, you want to perform the recalibration/export/print of the results after reviewing the chromatograms).
- Review the chromatograms: check the integration, etc.
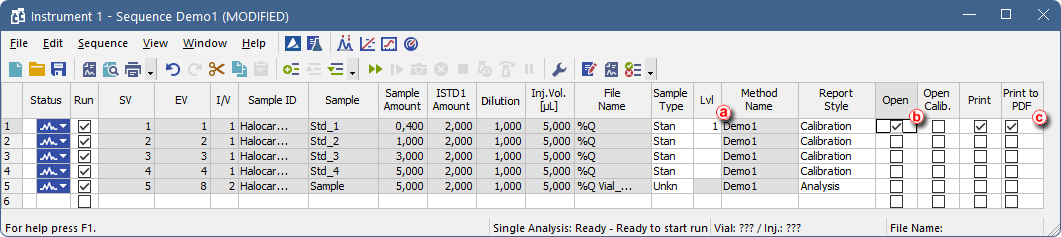
- Go to the measured sequence, set Levelsⓐ for calibration standards and enable Post Run options you wish to perform, e.g., Print To PDF ⓒ. It is also possible to uncheck (check) the Run checkbox at rows you don’t want to reprocess.
Note:
To print or export the data the chromatogram has to be opened in the Chromatogram window - this is achieved by checkbox in the Open ⓑ column on a given row.
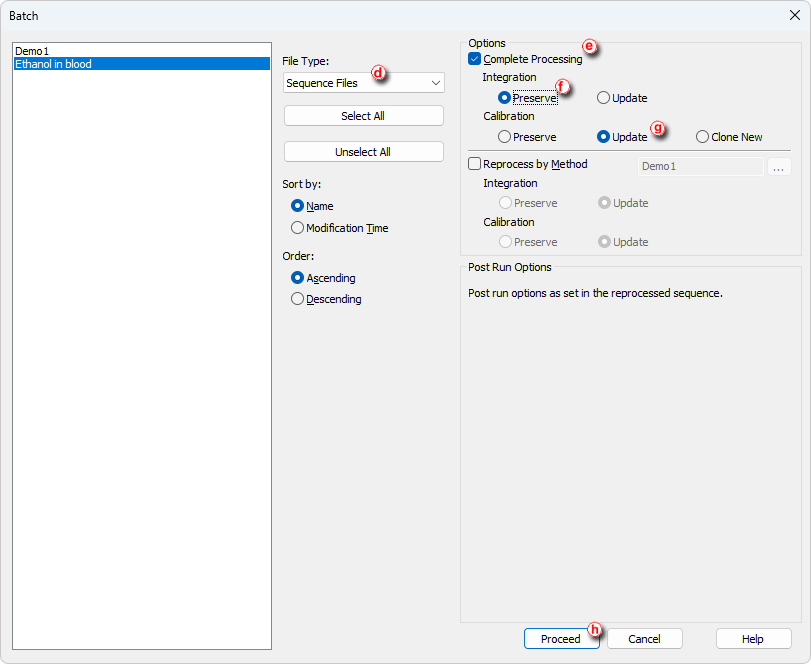
- Open the Batch dialog by using Analysis - Batch in the Instrument window.
- Switch File Type to Sequence Files ⓓ.
- Select the Sequence you wish to reprocess.
- Check the Complete Processing ⓔ checkbox.
- To preserve manually updated integration (this mainly considers small individual adjustments where optimized parameters weren't satisfactory) select the Preserve ⓕ in the Integrationpart of Options section.
- To perform recalibration based on updated integration select Update ⓖ in the Calibration part of Options section.
- Proceed ⓗ with the batch processing.
The recalibration will be performed and Post Run options defined in the sequence will be performed.
Caution:
The operations during Batch reprocessing are done row after row, injection after injection. In some situations it is necessary to perform the Complete Processing in two steps - first just the recalibrations, second the post-run actions (either using the Complete Processing again with the Integration a Calibration settings in the Options section to Preserve, or through running the post-process on selected chromatograms only through procedure described in Performing Post Run Options from Batch dialog section). For example, if the sequence is using calibration bracketing the unknowns are measured before second standards set and if the unknowns were reported during the recalibration step, the responses from the second standards set would be missing in the report.